The way DNA assembles itself inspires the creation of other structures that self assemble, says S.Ananthanarayanan.
The function of the DNA molecule is to help form a specific sequence of amino acids, each one picked out of the twenty amino acids that there are in all. Each such sequence is a separate protein, and the DNA, in a chain that can be thousands of units long, spells out millions of proteins and as many characteristics of living cells. But along with this function, one strand, or half a DNA molecule, after cell division, has the ability to assemble its complementary strand alongside itself, to create a complete molecule.
While there has been progress in using laboratory-made DNA sequences to create protein-like molecules ‘on order’, the so called ‘DNA Origami’, it is tempting to extend the self replicating property of DNA to other structures. This could get the structures to attach to specific complementary molecules and form sheets or 3-dimensional arrays. One possibility is with short sequences of amino acids, which are called peptides, and which connect to each other to form proteins. Or Berger, Lihi Adler-Abramovich, Michal Levy-Sakin, Assaf Grunwald, Yael Liebes-Peer, Mor Bachar, Ludmila Buzhansky, Estelle Mossou, V. Trevor Forsyth, Tal Schwartz, Yuval Ebenstein, Felix Frolow, Linda J. W. Shimon, Fernando Patolsky and Ehud Gazit of Tel Aviv University, the Institut Laue–Langevin, in Grenoble, Keele University in the UK and the Weizmann Institute of Science, Israel, report in the journal, Nature Nanotechnology, that they have got pairs of amino acid molecules to self-assemble into stable structures, which also showed interesting optical properties.

DNA and peptides
DNA – de-oxy-ribo-Nucleic Acid – is a long chain molecule, built around a backbone, to which are attached a series of chemical groups, called ‘bases’. There are only four kinds of bases, and these are adenine (A), guanine (G), thymine (T) and cytosine (C). Every group of three consecutive bases forms a ‘triad’ and each triad is the code for a particular amino acid. A DNA strand of nine bases would thus have three triads and code for three amino acids, connected in that order, although real DNA consists of hundreds of thousands of bases. Now, as each base can be or four kinds, a sequence of three bases can be formed in 4x4x4=64 ways. But a many of these ways code for the same, important amino acids, so that an error in one of the bases would leave the meaning of the triad unchanged, and only twenty amino acids, plus a signal for the start and the end of a sequence, are actually coded for.
As we now have means to synthesise the components of DNA and also ways to cut and splice segments of existing DNA, we have the ability to ‘create’ DNA sequences and this ability can be applied to generate large quantities of tailor-made organic chemicals, with the help of the DNA template.
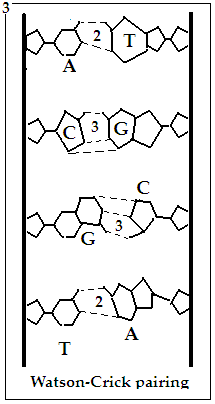
The other feature of the DNA molecule is that it consists of a pair of strands. While one strand has bases, A, G, T or C, attached along its length, the bases that appear on the complementary strand depend exactly on what bases there are in the first strand. Thus, where there is A, it would pair with T, and vice versa and where there is G, it would pair with C, and vice versa. At the time of cell division, the DNA molecule splits into two, both portions being complements of each other and in the new cells, the lone strands are able to assemble the second strand by controlling the order of A,T, G,C according to the T, A, C, G in the first strand.
The way this pairing happens is that A and T have a pair of hydrogen bonds each, with which they can connect, while G and C have three hydrogen bonds each and G and C hence form a couple. This architecture makes sure that a dangling A, for instance would only link to a T and if the base next to the A were a G, then the base next to the T would have to be a C, and so on.
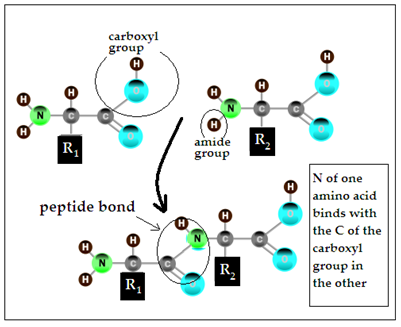
The other kind of specific linking is of the amino acids that the triads code for. Amino acids are so called because they consist of an amine, or nitrogen containing portion and an acid portion. The amine side ends with -NH2, while the acid portion ends with –COOH. In the peptide bond, the nitrogen of one amino acid binds with the carbon in the COOH portion of the second amino acid. The amine portion of the second amino acid can then bind with the COOH portion of a third amino acid, and so on.
In this way, the DNA, which replicates with the help of just four building blocks and the A-T and G-C pairing, called the Watson-Crick base pairing, generates a tool kit of just twenty amino acids, which can couple among themselves with versatility and stability. And these twenty four entities bring about the stupendous diversity of living things.
Self assembly
While structures that use peptides, or the combination of amino acids, are stable and can be versatile and flexible, they do not have the selectivity of the base pairing mechanism of DNA. There was hence an effort to combine the strengths of the two architectures, and this led to the idea of the PNA – which is not a Deoxyribo Nucleic Acid but a Peptide Nucleic Acid. The PNA, the name is a misnomer as it is not an acid, is a chain of amino acids, like glycine, the simplest, linked by peptide bonds, so that it forms a peptide backbone and has nucleobases (viz, A, T, G, C) as side chains. Such a structure could then show both the stability of the peptide as well as the possibility of Watson-Crick base pairing. One of the simpler peptides, called diphenylanailine, consisting of only two amino acids, is known to self-assemble into tubular nanostructures, which are stable, in fact show metal-like rigidity, withstand thermal and chemical stress and even display optical and piezoelectric properties. To better understand the mechanism of how simple peptides may combine, the Tel Aviv based group of scientists examined how all sixteen of the simplest, two-unit-PNAs could self-assemble in solution.
The sixteen di-PNAs are AA, AC, AG, AT, CA, CC, CG, CT, GA, GC, GG, GT, TA, TC, TG and TT. The group synthesised these di-PNAS and assayed for favourable conditions for self-organisation in different solvents. Assembly was found to arise in three of the sixteen di-PNAs – CG, GC and GG - all three containing guanine. In these cases, well-organized architectures, including long rods (tens of micrometres long) for CG and GC and spheroids with a diameter of 2–3 micrometres for GG were seen. These observations have pointed to the conditions, like the number of hydrogen bonds, which are necessary for structures of di-PNAS to be stable. And the fact of guanine, which is a key component in many natural nucleic acid structures, being present has led the team to more conclusions
Study of crystals of the structures, using X-Ray scattering, revealed that cytosine and guanine, with Watson-Crick type hydrogen bonding, led to stacking interactions between molecules. It is hence genuinely a case of DNA style selection brought to play to create stable structures built from peptides. It was also discovered that the structures displayed fluorescence, or the absorption and emission of light, with features, like wide span of wavelengths, that make the structures suitable for use as organic light emitting material in optoelectronics, optical communication, display, etc.
------------------------------------------------------------------------------------------